
UTGB Toolkit - BED Track
BED track is for displaying UCSC's BED format data. To create a BED track, add the following XML fragment to your (project folder)/src/main/webapp/view/default-view.xml file (see also How to customize browser contents). Do not foget to set your BED file path to the fileName property:
<track className="org.utgenome.gwt.utgb.client.track.lib.BEDTrack" name="BED Track" pack="true">
<property key="type">image</property>
<property key="leftMargin">100</property>
<property key="trackBaseURL">utgb-core/BEDViewer</property>
<!-- this track reads BED data from db/sample.bed -->
<property key="fileName">db/sample.bed</property>
</track>Since utgb-shell-1.2.4, utgb import commands is available for creating database indexes (SQLite DB) of BED data. To create an SQLite index of your BED data, say db/reads.bed, type the following commands:
> utgb import -t bed db/reads.bedThe SQLite index will be generated as db/reads.bed.sqlite file. When some SQLite DB file (suffixed with .sqlite) is found for the BED data, BEDTrack issues database queries to reads.bed.sqlite file, instead of reading reads.bed file.
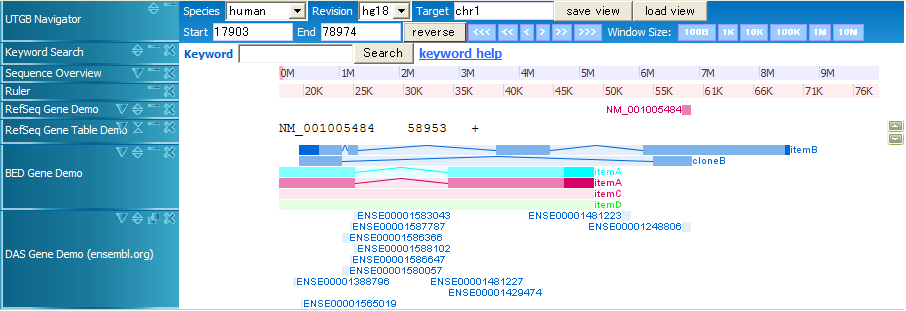
db/sample.bed
browser position chr1:1-100000 browser hide all track name="Item,RGB,Demo2" description="Item RGB demonstration2" visibility=2 itemRgb="On" useScore=1 color=0,128,0 url="http://genome.ucsc.edu/goldenPath/help/clones.html#$$" chr1 20000 70000 itemB 200 - 22000 69500 0 4 4330,1000,5500,15000 0,5000,20000,35000 chr1 20000 60000 cloneB 900 - 20000 60000 0 2 4330,3990, 0,36010 chr1 10000 50000 itemA 960 + 11000 47000 0,255,255 2 15670,14880, 0,25120 chr1 10000 50000 itemA 960 + 11000 47000 0 2 15670,14880, 0,25120 chr1 10000 50000 itemC 960 + 11000 47000 0 chr1 10000 50000 itemD 960 + 11000 47000 0,255,0 chr22 20100000 20100100 first chr22 20100200 20100300 second chr22 20100400 20100500 thirds chr7 127472363 127473530 Pos2 200 chr7 127472363 127473530 Pos2 200 + chr7 127472363 127473530 Pos2 200 + 127472363 127473530 chr7 127472363 127473530 Pos2 200 + 127472363 127473530 255,0,0