
In the following explanation, we assume that the project folder name is demo.
To create a genome browser interface, type utgb gwt in the project folder:
leo@lecture-2008/demo> utgb gwt
[ScaffoldGenerator] output folder: .
[ScaffoldGenerator] packageName = demo
[ScaffoldGenerator] projectName = demo
[ScaffoldGenerator] moduleName = demo.gwt.UTGBEntry
[ScaffoldGenerator] clientPackageName = demo.gwt.client
[ScaffoldGenerator] entryPoint = demo.gwt.client.UTGBEntry
[ScaffoldGenerator] create a directory: ./src/main/java/demo/gwt
[ScaffoldGenerator] create a directory: ./src/main/java/demo/gwt/public
[ScaffoldGenerator] create a directory: ./src/main/java/demo/gwt/client
[ScaffoldGenerator] create a file: ./src/main/java/demo/gwt/public/UTGBEntry.html
[ScaffoldGenerator] create a file: ./src/main/java/demo/gwt/UTGBEntry.gwt.xml
[ScaffoldGenerator] create a file: ./src/main/java/demo/gwt/client/UTGBEntry.java
[UTGBShell] create a file: ./eclipse/demo-gwt.launch
[UTGBShell] create a file: ./eclipse/demo-gwt-mac.launchThe following steps compile the genome browser interface:
leo@lecture-2008/demo> utgb compilewhich converts Java codes into JavaScript codes. This compile process usually takes 30 seconds to 1 minute.
After the compilation finished, launch the server:
leo@lecture-2008/demo> utgb server -gThen, open the url http://localhost:8989/demo/UTGBEntry.html with your web browser.
utgb eclipse command generates Eclipse project files, .project, .classpath into the project folder:
leo@lecture-2008/demo> utgb eclipseThen import the demo folder as a project in Eclipse. See also this document for the other necessary steps to use UTGB Toolkit with Eclipse. At least you have to set M2_REPO and UTGB_HOME variables in Eclipse.
In the Debug Configurations menu in Eclipse, you will find demo-gwt, demo-gwt-mac, demo-server Java application launch configurations.
The browser interface of UTGB consists of set of tracks. To change the tracks displayed in the browser, we call this set of tracks track view, you have to edit (project folder)/src/main/webapp/view/default-view.xml file or add another view XML file, e.g., (project folder)/src/main/webapp/view/custom-view.xml, etc. The defalut-view.xml file is loaded when opening the browser. To change the view file to be loaded at the startup time, use the following URL query parameter:
http://localhost:8989/UTGBEntry.html?view=custom-view.xmlThe genome browser will automatically load src/main/webapp/view/custom-view.xml file.
Here is the configuration setting when developing with Eclipse:
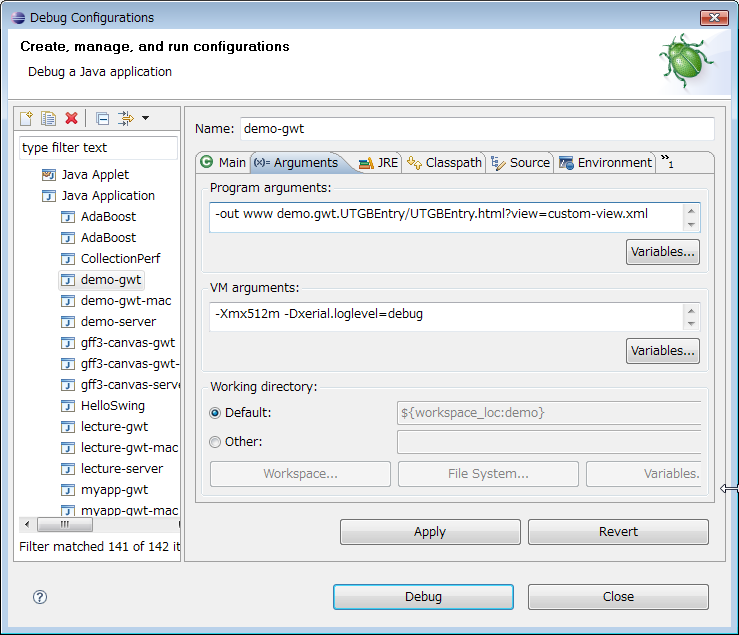
The view XML file defines track contents to be displayed on the browser.
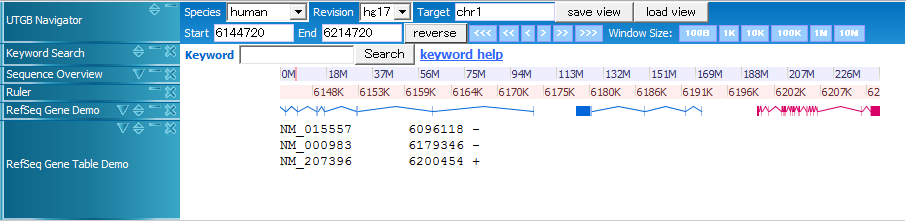
<trackGroup className="org.utgenome.gwt.utgb.client.track.TrackGroup" name="human">
<property key="name">human</property>
<!-- initial display location -->
<groupProperties>
<property key="revision">hg17</property>
<property key="species">human</property>
<property key="target">chr1</property>
<trackWindow start="6144720" end="6214720" width="700"/>
</groupProperties>
<!-- UTGB Navigator configurations -->
<track className="org.utgenome.gwt.utgb.client.track.lib.NavigatorTrack" name="UTGB Navigator" height="41" pack="true">
<!-- menu contents -->
<property key="sequenceList">[{"species":"human", "revision":["hg18","hg17"]},{"species":"mouse", "revision":["mm9"]}, {"species":"drosophila", "revision":["dm3"]}]</property>
</track>
<!-- keyword search track provides links to other locations on genomes -->
<track className="org.utgenome.gwt.utgb.client.track.lib.KeywordSearchTrack" name="Keyword Search" pack="true">
<property key="keyword.examples">["Scaffold number (e.g. scaffold123)",
"GenBank accession number (e.g. BJ013363)",
"UniGene cluster identifier (e.g. Ola.5710)",
"Clone number (e.g. MF01SSA159G06)"]
</property>
<property key="species.scope">any</property>
</track>
<!-- ruler for the whole sequence -->
<track className="org.utgenome.gwt.utgb.client.track.lib.SequenceRulerTrack" name="Sequence Overview" height="14" pack="true">
<property key="leftMargin">100</property>
</track>
<!-- ruler for the track -->
<track className="org.utgenome.gwt.utgb.client.track.lib.RulerTrack" name="Ruler" height="14" pack="true">
<property key="leftMargin">100</property>
</track>
<!-- Refseq gene track provided as a PNG image -->
<track className="org.utgenome.gwt.utgb.client.track.lib.GenomeTrack" name="RefSeq Gene Demo" pack="true">
<!-- use an image data as a track content -->
<property key="type">image</property>
<!-- if you have to open spaces in the left-side set this value -->
<property key="leftMargin">100</property>
<!-- this track retrieves the contents from (server_URL)/utgb-core/GeneViewer -->
<property key="trackBaseURL">utgb-core/GeneViewer</property>
</track>
<!-- Refseq gene table track provided as a text data -->
<track className="org.utgenome.gwt.utgb.client.track.lib.GenomeTrack" name="RefSeq Gene Table Demo" height="150" pack="false">
<!-- use an iframe html element as a track content -->
<property key="type">frame</property>
<property key="leftMargin">100</property>
<!-- this track retrieves the contents from (server_URL)/utgb-core/GeneViewer.tab -->
<property key="trackBaseURL">utgb-core/GeneViewer.tab</property>
</track>
<!-- you can add other tracks here -->
</trackGroup>
To make the change on the view XML file effective, you have to close the utgb server, and do 'utgb server' again.