
Currently, UTGB toolkit provides three ways to develop you own genome tracks:
Using a genome track for image data is the simplest method to visualize genome-related data in your UTGB browser. Given a trackBaseURL value, this track generates an URL consisting of
(trackBaseURL)?group=utgb&species=..&revision=..&name=..&start=..&end=..&width=..The above URL query parameters (..) are generated from the current displayed position in the UTGB browser. The generated URL must produce some image data using these parameter values.
To display genome track on your genome browser, you have to add the following XML fragments. The type value in the property tag must be image, and the content of the track is retrieved from the URL generated from the given trackBaseURL + track group properties (%q symbol will be replaced to the URL query paramters).
<track className="org.utgenome.gwt.utgb.client.track.lib.GenomeTrack"
name="RefSeq Gene Demo" pack="true">
<property key="type">image</property>
<property key="leftMargin">100</property>
<property key="trackBaseURL">utgb-core/GeneViewer?%q</property>
</track>For the local server, the above trackBaseURL value corresponds to http://localhost:8989/myapp/utgb-core/GeneViewer. You can use full-length URL name beginning with http:// to specify arbitrary web pages.
This code recieves parameter values, species, revision, parameters, etc. from the genome browser interface (client-side code), then the server-side code retrieves RefSeq gene data from the http://utgenome.org/api/refseq/ API. Next, the GeneViewer draws the graphics of RefSeq genes within the specified region. If the accessed URL contains a suffix .tab, i.e. utgb-core/GeneViewer.tab, it simply returns the data of retrived genes in a text format.
To draw graphics, this web action code uses GeneCanvas class, the source code of which is available from http://svn.utgenome.org/utgb/trunk/utgb/utgb-core/src/main/java/org/utgenome/graphics/GeneCanvas.java
/**
* Gene Viewer
*/
public class GeneViewer extends WebTrackBase {
private static final long serialVersionUID = 1L;
private static Logger _logger = Logger.getLogger(GeneViewer.class);
// public fields for receiving URL query parameters
public String species = "human";
public String revision = "hg18";
public String name = "chr1";
public long start = 1;
public long end = 1000000;
public int width = 800;
public GeneViewer() {
}
static class GeneList {
public List<Gene> gene = new ArrayList<Gene>();
}
public void handle(HttpServletRequest request, HttpServletResponse response)
throws ServletException, IOException {
String refseqURL =
String.format("http://utgenome.org/api/refseq/%s/%s/%s:%d-%d/list.json",
species, revision, name, start, end);
URL apiURL = new URL(refseqURL);
// retrieve gene data from the UTGB web API
GeneList geneList = Lens.loadJSON(GeneList.class, apiURL);
String actionSuffix = getActionSuffix(request);
if (actionSuffix.equals("tab")) {
// output gene data as text
response.setContentType("text/plain");
for (Gene each : geneRetriever.getResult()) {
response.getWriter().println(
String.format("%s\t%s\t%s",
each.getName(), each.getStart(), each.getStrand()));
}
}
else {
// draw graphics of the RefSeq genes
GeneCanvas geneCanvas = new GeneCanvas(width, 300, new GenomeWindow(start, end));
geneCanvas.draw(geneRetriever.getResult());
response.setContentType("image/png");
geneCanvas.toPNG(response.getOutputStream());
}
}
}This API uses the following URL pattenrs to retrieve RefSeq gene data:
http://utgenome.org/api/refseq/human/hg18/chr1:1-100000/list.json
This url returns JSON data as follows:
{"gene":
[{"name":"NM_001005484","end":59871,"start":58953,"strand":"+",
"exon":[{"end":59871,"start":58953}],
"cDS":[{"end":59871,"start":58953}]}]}This data structure corresponds to org.utgenome.gwt.utgb.client.bio.Gene class http://svn.utgenome.org/utgb/trunk/utgb/utgb-core/src/main/java/org/utgenome/client/bio/Gene.java. To learn how to bind the above JSON data to the Java object, see also Smart Data Binding in UTGB.
To embed arbitrary HTML contents as a track, e.g., web pages, web action for generating HTML dat etc., the iframe track is useful. The view XML format and URL to be accessed are the same with the image track, except the type property is frame.
-track
-class: GenomeTrack
-name: RefSeq Gene Table Demo
-height: 100
-pack: true
-properties
-type: frame
-trackBaseURL: utgb-core/GeneViewer.tabTo implement your own tracks using GWT, you have to understand how the UTGB displays track contents. A track group is a container of several tracks, and holds common variables that can be accessed from its contained tracks. For example, the track group variables for speciying a location on a genome sequence are species (e.g., human, medaka, etc.), revision (e.g, hg18, version1.0, etc.) and name (e.g., chr1, scaffold1, etc.). In the Java code of the UTGB tracks, PropertyReader(PropertyWriter) class can be used to read/write these variables, and changes to these variables will be notified to the other tracks in the same track group. To capture these events, you need to override onTrackGroupPropertyChange() method in the track implementation.
String species = getTrackGroup().getPropertyReader().getProperty("species");getTrackGroup().getPropertyWriter().setProperty("species", "human");The location in the currently displayed genome sequence can be accessed through TrackWindow class. The change to the TrackWindow also will be notified to the other tracks in the track group.
long startPosOnGenome = getTrackGroup().getTrackWindow().getStartOnGenome();
long endPosOnGenome = getTrackGroup().getTrackWindow().getEndOnGenome();Given track group properties, each track generates HTML contents to display. Track contents allowed to be arbitrary HTML data, including HTML pages on the web, server-generated image, text, database query results, etc.
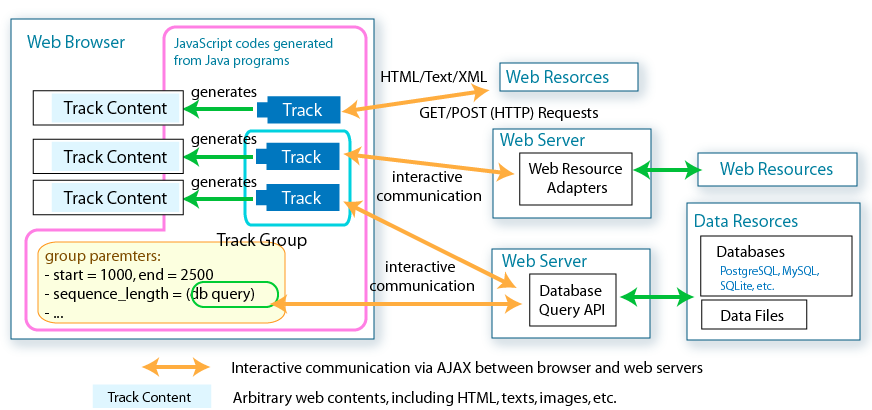
Google web toolkit enables developing interactive HTML user interfaces, which can be changed dynamically in response to mouse clicks, keyboard inputs, etc. Standard HTML components are wrapped as Widget classes in Java, so no need exists to write HTML tags.
Implementing custom tracks is the only way to dynamically change the track group properties, track window locations in response to the user input, such as mouse clicks, keybord inputs etc.
GWT code is a client side program that runs on the web browser (IE, Firefox, etc.) so it is impossible to access databases on the server through JDBC or object-data mapping functionality described in dbaccess.html. There are several limitation on available Java libraries in GWT. For details of GWT programming, see the follwing link: http://www.xerial.org/trac/Xerial/wiki/WebApplication
To take interactive commucations between server-side programs (e.g. web actions) and custom tracks (client-side program), for example, retrieving database contents and changing track contents according to the query result, you have to learn how to implement RPC (Remote Procedure Call), which enables invoking methods on a server-side program from the client-side code.
Track implementation
Extend TrackBase class, and add factory method (public static, and returns TrackFactory).
In the track code, you have to implement getWidget() method, which returns track content.
package demo.gwt.client;
import org.utgenome.gwt.utgb.client.track.Track;
import org.utgenome.gwt.utgb.client.track.TrackBase;
import org.utgenome.gwt.utgb.client.track.UTGBProperty;
import org.utgenome.gwt.widget.client.Style;
import com.google.gwt.user.client.ui.ClickListener;
import com.google.gwt.user.client.ui.HorizontalPanel;
import com.google.gwt.user.client.ui.Hyperlink;
import com.google.gwt.user.client.ui.Widget;
public class SampleTrack extends TrackBase
{
/**
* Creates a factory of this track
*
* @return
*/
public static TrackFactory factory()
{
return new TrackFactory() {
@Override
public Track newInstance()
{
return new SampleTrack();
}
};
}
private HorizontalPanel panel = new HorizontalPanel();
private Hyperlink humanLabel = new Hyperlink("human", "species=human");
private Hyperlink medakaLabel = new Hyperlink("medaka", "species=medaka");
public SampleTrack()
{
super("Sample Track");
// set design
Style.padding(humanLabel, 5);
Style.padding(medakaLabel, 5);
panel.add(humanLabel);
panel.add(medakaLabel);
humanLabel.addClickListener(new ClickListener() {
public void onClick(Widget arg0)
{
// change track group property
getTrackGroup().getPropertyWriter().setProperty(UTGBProperty.SPECIES, "human");
}
});
medakaLabel.addClickListener(new ClickListener() {
public void onClick(Widget arg0)
{
// change track group property
getTrackGroup().getPropertyWriter().setProperty(UTGBProperty.SPECIES, "medaka");
}
});
}
public Widget getWidget()
{
return panel;
}
}Add the TrackFactoryHolder.addTrackFactory() method to the main() method of your UTGBEntry.java code.
//--------------------------------------
// utgb-shell Project
//
// UTGBEntryPoint.java
// Since:
//
//--------------------------------------
package demo.gwt.client;
import org.utgenome.gwt.utgb.client.UTGBEntryPointBase;
import org.utgenome.gwt.utgb.client.track.TrackFactoryHolder;
/**
* Entry point of the UTGB Browser. Edit the following files to change the
* appearance of the browser: src/main/webapp/view/default-view.xml
* public/browser.html
*
*/
public class Browser extends UTGBEntryPointBase
{
@Override
public void main()
{
// register your custom track
TrackFactoryHolder.addTrackFactory
("demo.gwt.client.SampleTrack", demo.gwt.client.SampleTrack.factory());
// This line insert the track display in the <div id="utgb-main"></div> part
// You can defiene track contents to be displayed
// in the view XML file (src/main/webapp/view/default-view.xml is used in default)
displayTrackView();
// add your GWT codes here
}
}<track className="demo.gwt.client.SampleTrack" name="Sample" pack="true">
</track>Add the above XML fragment to your view XML file.
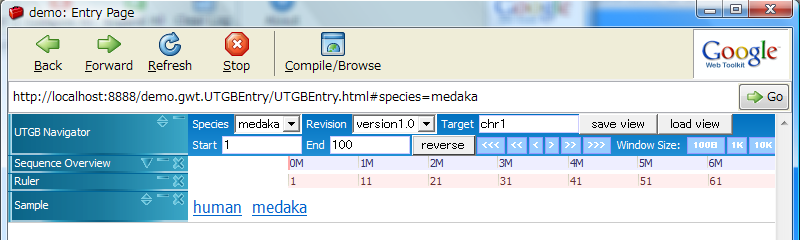
Clicking on the human or medaka labels changes the track group properties.
If you override saveProperties() and restoreProperties() method, you can pass several parameter values to the track:
int leftMargin = 0;
@Override
public void saveProperties(Properties saveData)
{
// used to save property value to the view XML file
saveData.add("leftMargin", leftMargin);
}
@Override
public void restoreProperties(Properties properties)
{
// load the leftMargin value from the view XML file
leftMargin = properties.getInt("leftMargin", leftMargin);
}<track className="demo.gwt.client.SampleTrack" name="Sample" pack="true">
<property key="leftMargin">50</property>
</track>